What is Xenium?
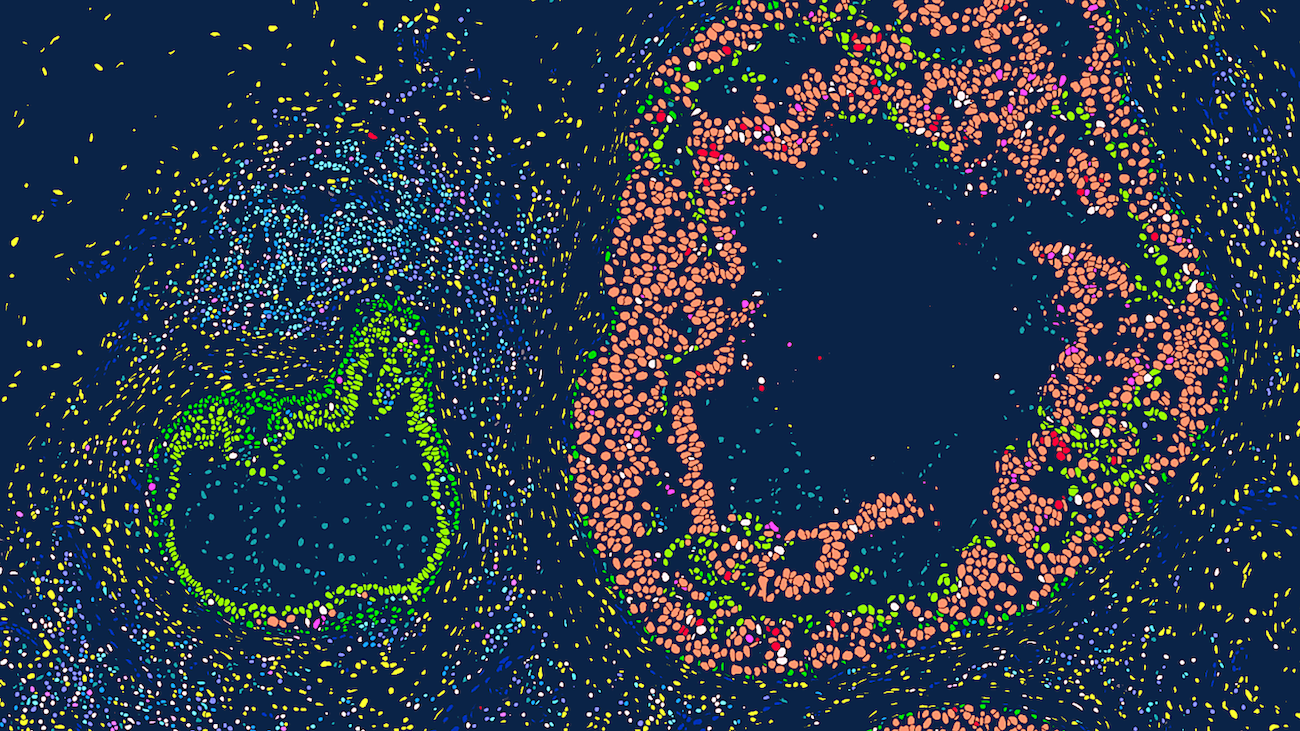
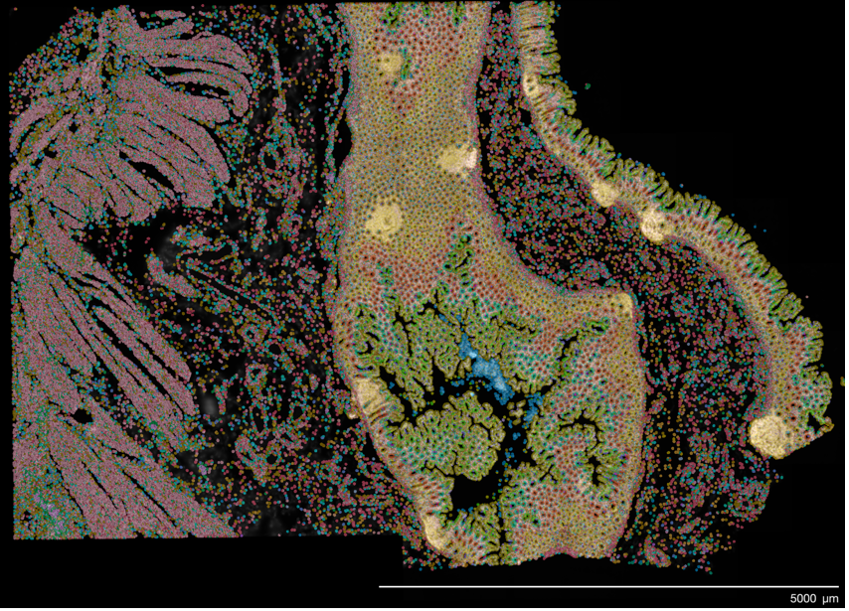
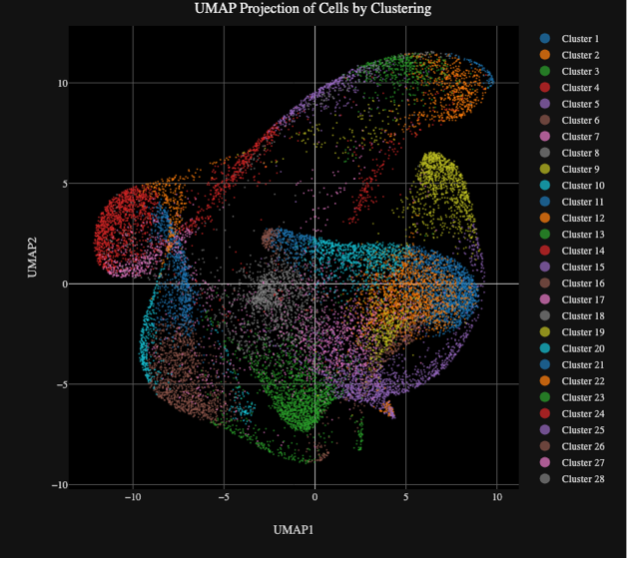
Xenium is a high-resolution, imaging-based in situ spatial profiling technology from 10x Genomics that allows for simultaneous expression analysis of RNA targets (currently in range of 100’s) within the same tissue section. This assay can identify the location of target transcripts within the tissue, providing a single cell resolution map of expression patterns of all genes that are included in the selected probe panel. Researchers can choose between off-the-shelf panels of genes designed to identify cells types and interrogate specific phenotypes, add on 50-gene or 100-gene panels to these off-the-shelf panels, or design a full custom standalone panel to specifically tailor for their tissue and research questions. test
Researchers can utilize a multimodal segmentation panel to enhance cell segmentation accuracy by incorporating additional morphology markers. 5,000-plex target panels (Xenium Prime Assay) are now available for both mouse and human, alongside existing tissue-specific and custom-designed panel options (Xenium v1 Assay). Data is easily viewable using Xenium Explorer, 10x Genomics’ dedicated desktop visualization tool, and is compatible with a rapidly expanding ecosystem of third-party downstream analysis tools for spatial transcriptomics.
Key Features
- Single-Cell Resolution: Spatial information at the single-cell level post-segmentation.
- Sensitive and Accurate Chemistry: Padlock probe and rolling circle amplification promote greater specificity and allow better sensitivity.
- Speed: Efficient instrument run time and onboard image data processing provide viewable data quickly.
How Does it Work?
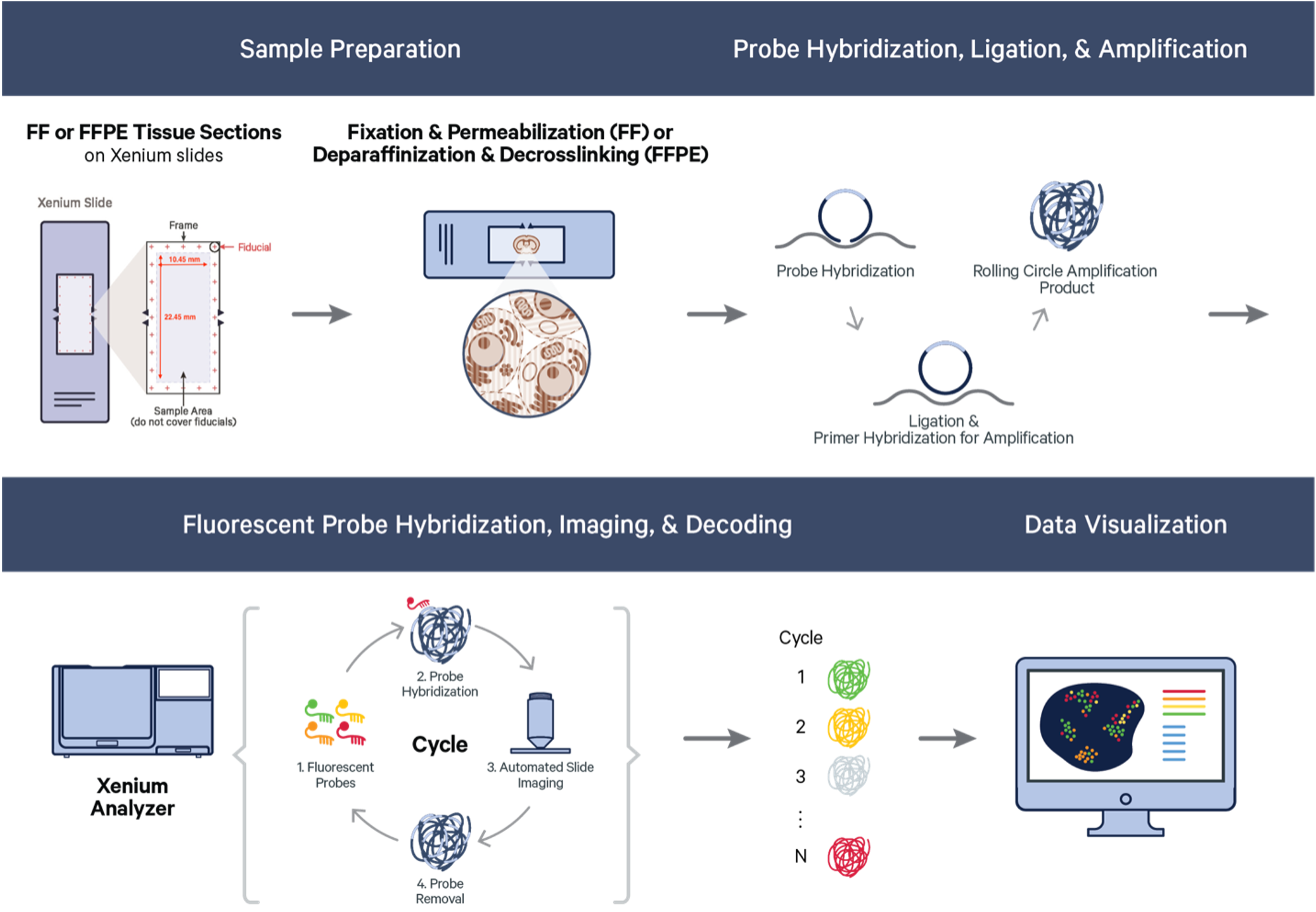
Xenium starts with FFPE or fresh frozen sections on specific Xenium slides. The section tissue is processed and selected probes hybridized to the RNA targets within the tissue. Bound probes are ligated, forming a circular template that allows rolling circle amplification of the probe and a unique barcode. On the Xenium platform, the tissue with amplified probe products are sequenced through a series of cycles and onboard image data processing that allow identification and location of each transcript target. Resulting data is immediately viewable and is compatible with a easy to use data viewer and with many third-party downstream analysi tools, which continue to rapidly develop.
Project Support
To get more information or to discuss a potential supported project at CCR, please contact the Single-Cell and Spatial Core (SCSC).
Disclaimer: The information provided here is based on the latest available data and may be subject to updates.