The OSTR offers cutting-edge technology platforms to the CCR scientific community through centralized facilities. The videos accessed through this page are designed to introduce the various scientific methodologies OSTR makes available through the cores on both Frederick and Bethesda campuses.
Spatial proteomics with the Imaging Mass Cytometry
Milind M. Pore, Ph.D. (FNLCR), Scientist II, Mass Cytometry Core (MCC), Cancer Research Technology Program.
Share Video
Seminar
Empowering Biological Discovery Through Computational Chemistry and Molecular Modeling
This seminar in this series features two presentations from Lalith Perera, Ph.D. and Robin Evans Stanley, Ph.D.
Lalith Perera, Ph.D.
Share Video
Core Spotlight
Biomodal’s 6-Base Genome to Decode Health and Disease
Experience richer, faster reads of genetic and epigenetic information with the 6-base genome. Mark Consugar, MS
Associate Director – Scientific Affairs.
Share Video
Seminar
CCR Volume Electron Microscopy (CVEM) Core
This seminar in this series features three presentations from Kedar Narayan, Ph.D., Daniel Castranova, M.S., and Quanlong Lu, Ph.D.
Share Video
Core Spotlight
NIH Mouse Imaging Facility (MIF)
NIH Intramural Core Spotlight Seminar: NIH Mouse Imaging Facility (MIF)
Share Video
Core Spotlight
Functional Genomics Lab
Ken Cheng, Ph.D., Director, Functional Genomics Lab, NCATS. Presentation Title: “Overview of the Functional Genomics Laboratory”.
Share Video
Core Spotlight
NHGRI Zebrafish Core
Raman Sood, Ph.D., Associate Investigator, Office of Scientific Core Facilities, Director, Zebrafish Core, NHGRI. Presentation Title: “Genome editing services and phenotyping resources provided by the NHGRI Zebrafish Core”.
Share Video
Core Spotlight
NIAID’s Mouse Genetics and Gene Modification Section and Laboratory of Immune Systems Biology (LISB)
CRISPR/Cas9 genome edited mouse models: Gene Knockout, Knock-In, Conditional Knockout and more.
Share Video
Core Spotlight
Why Cryo-Electron Microscopy?
Capturing High Resolution Images of Macromolecular Complexes.
Share Video
Educational
Trans-NIH Metabolomics Core and NIEHS Metabolomics Core Facility
“Shared Chemistries of Disease: Hypothesis-driven metabolomic and lipidomic applications across disease systems.
Share Video
Core Spotlight
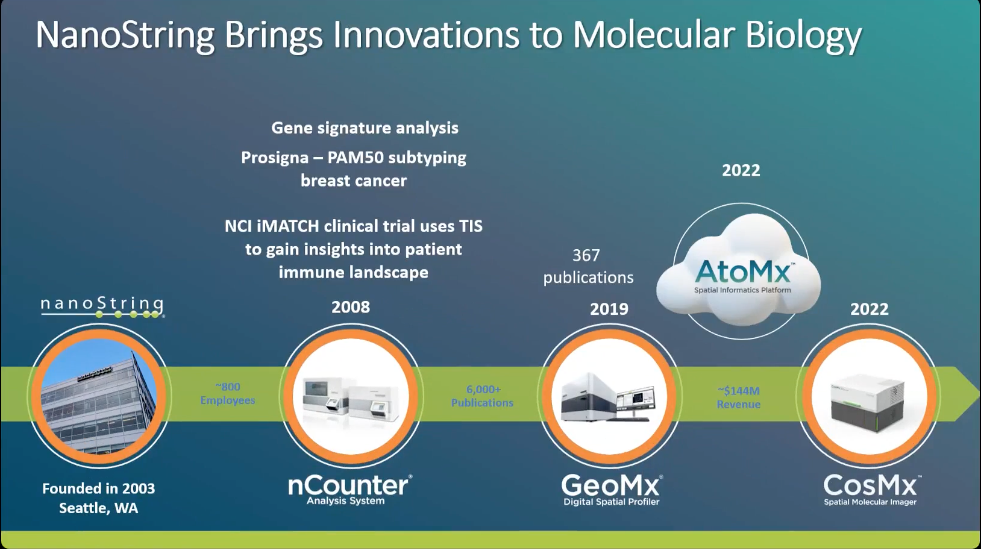
Nanostring Brings Innovation to Molecular Biology
Profile the entire transcriptome and more than 570 proteins on a single slide with the GeoMx Digital Spatial Profiler.
Share Video
Seminar
Nanopore Sequencing for Single cell and Spatial Applications
Oxford Nanopore Technologies and 10X Genomics provide updates on using long read sequencing for single cell and spatial applications.
Share Video
Seminar
BEPS MAI Unit
Heather Kalish, Ph.D., Unit Chief, Micro Analytical Immunochemistry Unit, BEPS, NIBIB. “Microanalytical Immunochemistry- what we do and how we can help!”.
Share Video
Core Spotlight
Proteomics Analysis using Mass Spectrometry
Analysis of Post-Translational Modifications by Mass Spectrometry.
Share Video
Educational
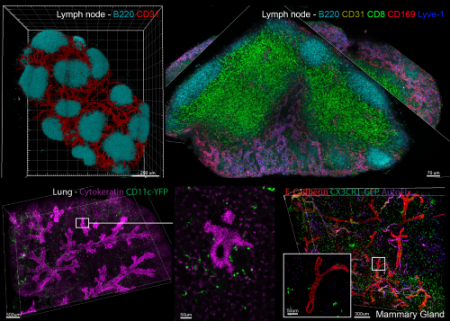
Center for Advanced Tissue Imaging (CAT-I)
This seminar will detail IBEX, RAPID, Ce3D, and other technologies, show examples of their application to various tissues and cancers.
Share Video
Seminar
Crosslinking and Limited Proteolysis
Structural Mass Spectrometry related to crosslinking and limited proteolysis mass spectrometry approaches.
Share Video
Educational
Animal Research Services
NIH ORS and NIH CREx Panel Discussion Series. Panel 2: Animal Model Research Services. February 24, 2022.
Share Video
Core Spotlight
Structural Mass Spectrometry
Structural Mass Spectrometry related to crosslinking and limited proteolysis mass spectrometry approaches.
Share Video
Educational
10X Platforms
Pushing the Boundaries of Single Cell with Chromium 10X.
Share Video
Seminar
Core Spotlight Seminar
“Biology Goes 3D: Volume Electron Microscopy of Biological Specimens”.
Share Video
Core Spotlight
Core Spotlight Seminar
Mitochondrial Network Formation as a Tissue-Specific Phenomenon in a Mammalian Model of Extreme Metabolism.
Share Video
Core Spotlight
Quantitative Mass Spectrometry
Quantitative mass spectrometry, specifically the global discoveryexperiments in mass spectrometry.
Share Video
Educational
NINDS Viral Production Core
“An Introduction to Viral Vectors and the NINDS Viral Production Core Facility”.
Share Video
Core Spotlight
PTM Mass Spectrometry
PTM mass spectrometry, specifically the analysis of post-translational modifications by mass spectrometry.
Share Video
Educational
NINDS Laboratory of Functional and Molecular Imaging
“Viral Enabled Study of the Neurons and Circuits Underlying the Brain’s Response to Peripheral Injury”.
Share Video
Core Spotlight
Interactome Mass Spectrometry
Interactome mass spectrometry approaches specifically, proximity-dependent biotinylation for mapping protein complexes mass spectrometry.
Share Video
Educational
Core Services: Mass Spectrometry
NIH ORS and NIH Collaborative Research Exchange (CREx) Panel Discussion Series. Panel 1. October 27, 2021.
Share Video
Core Spotlight
Visualizing Cells in 3-D and at the Nanoscale Level
An introduction to focused ion beam scanning electron microscopy (FIB-SEM) as it relates to visualizing cells in 3-D and at the nanoscale level.
Share Video
Educational