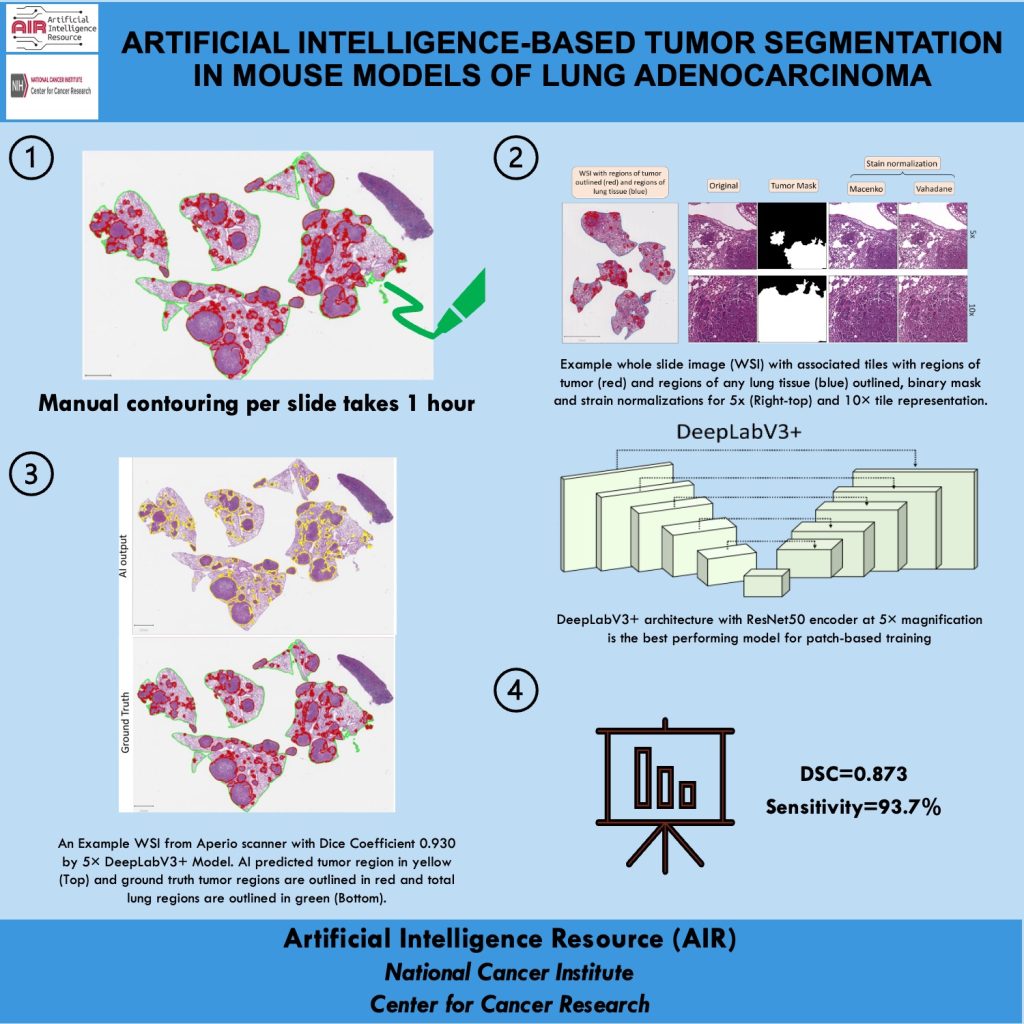
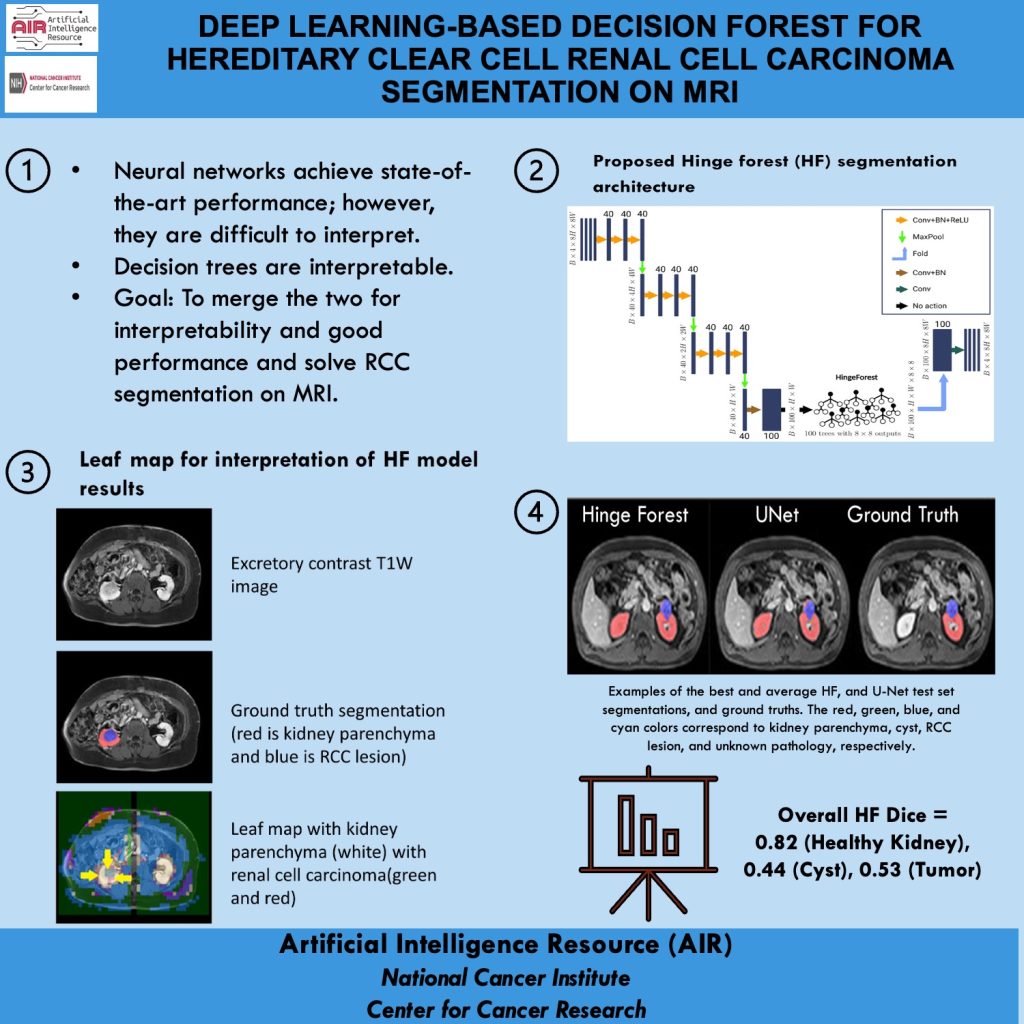
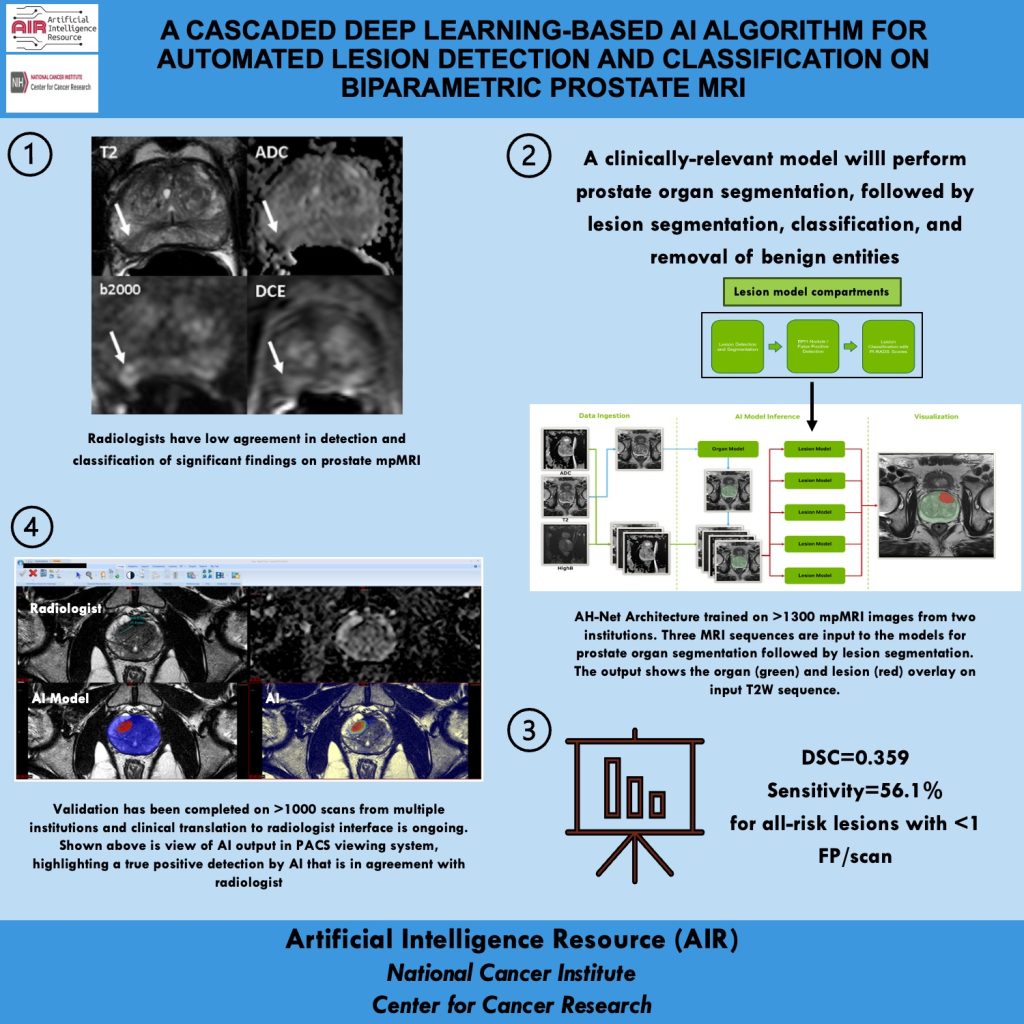
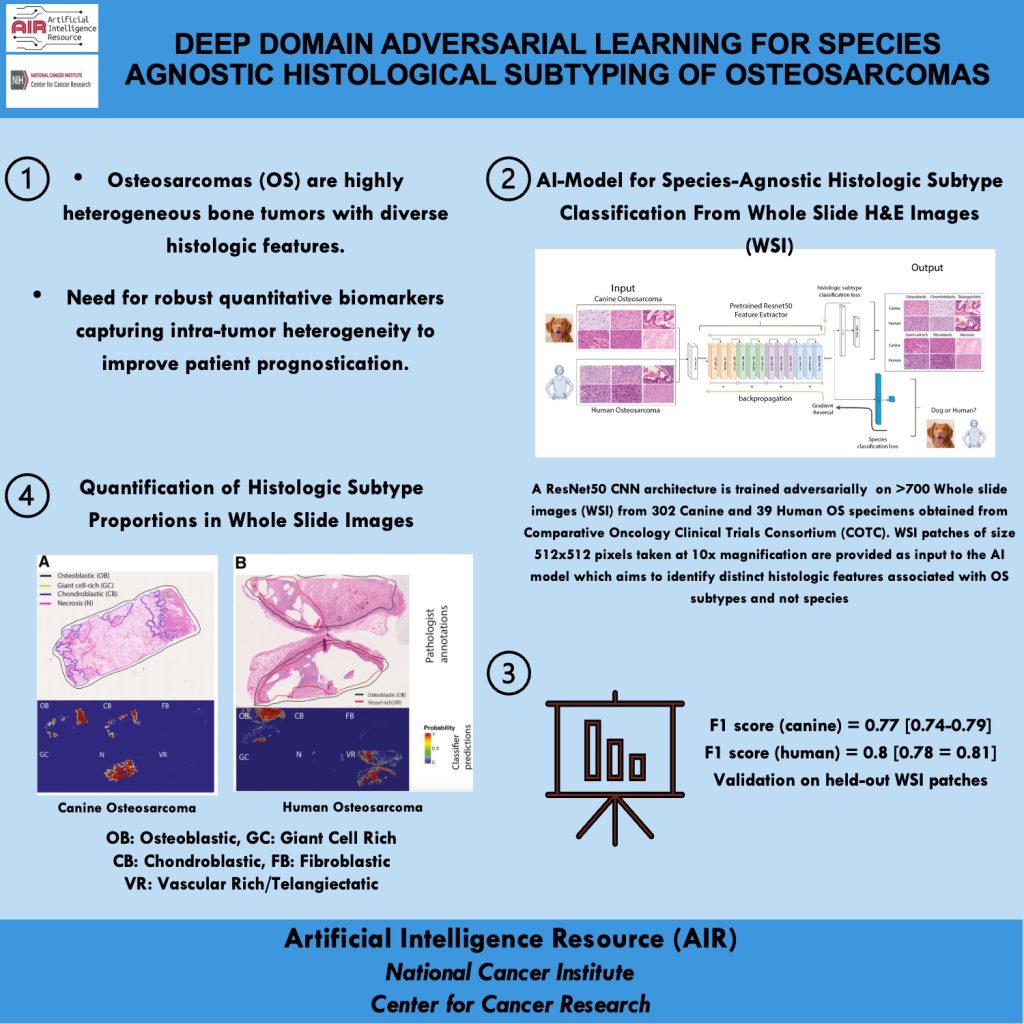
Mission:
The goal of Artificial Intelligence Resource (AIR) is to make AI tools available to Center for Cancer Research (CCR) investigators. The strength of AI is that algorithms can be trained to seek specific information that may be scientifically or clinically important. AIR will mainly focus on “Computer Vision” which analyzes medical images, such as radiologic, digital pathology, video/endoscopy, and optical imaging among others. Examples of potential projects include developing better screening, detection methods or predictive markers, or improving procedures among many others. Both clinical and laboratory-based imaging projects will be considered. Please refer to our ongoing Projects and Prior Publications for more information.